[ad_1]
Summary: Study details a signaling pathway that prevents the overproduction of snRNPs when they are not required.
Source: University of Würzburg
Neurodegenerative diseases, such as various forms of senile dementia or amyotrophic lateral sclerosis (ALS), have one thing in common: large amounts of certain RNA-protein complexes (snRNPs) are produced and deposited in the nerve cells of those affected – and this hinders the function of the cells. The overproduction is possibly caused by a malfunction in the assembly of the protein complexes.
How the production of these protein complexes is regulated was unknown until now. Researchers from Martinsried and Würzburg in Bavaria, Germany, have solved the puzzle and now report on it in the open access journal Nature Communications.
They describe in detail a signaling pathway that prevents the overproduction of snRNPs when they are not needed. The results should make it possible to better understand the processes in motor neuron diseases and senile dementia.
The research group led by Professor Michael Sendtner and Dr. Michael Briese from the Institute of Clinical Neurobiology at Julius-Maximilians-Universität Würzburg (JMU) was in charge of the publication. Professor Utz Fischer and Pradhipa Ramanathan from the JMU Institute of Biochemistry were also involved, as was a team from the Max Planck Institute of Biochemistry in Martinsried.
The next steps in research
Further investigations shall now show how the synthesis and degradation of excess snRNPs are regulated in nerve cells. The scientists hope that in the end they will be able to identify new therapeutic options for neurodegenerative diseases.
About this neuroscience research news
Source: University of Würzburg
Contact: Michael Sendtner – University of Würzburg
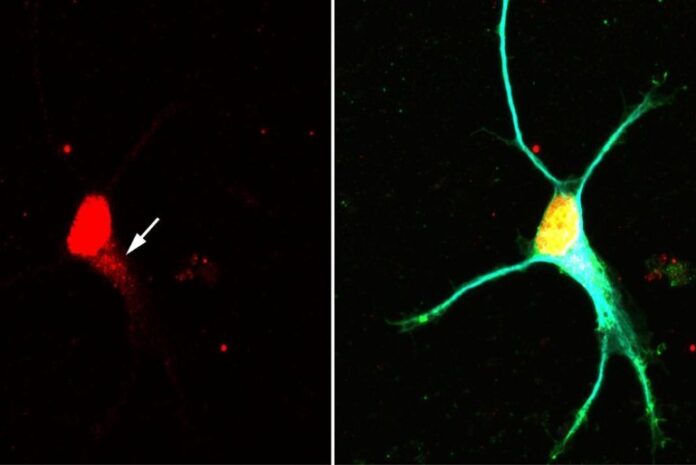
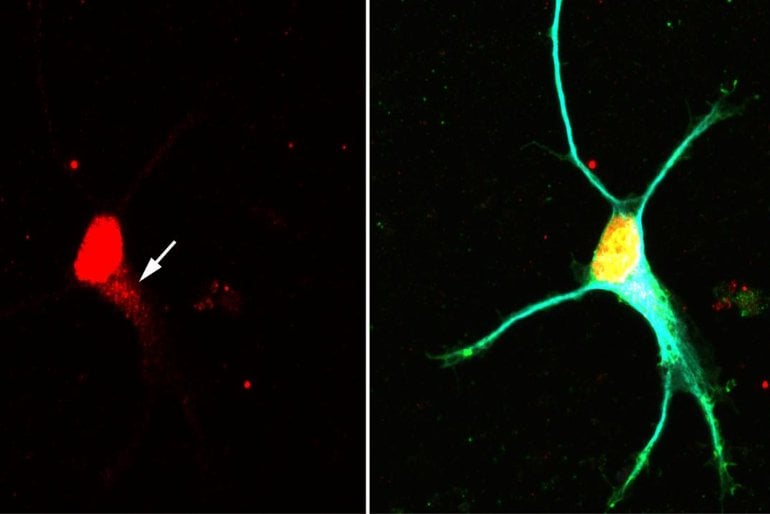
Image: The image is credited to Changhe Ji / University of Würzburg
Original Research: Open access.
“Interaction of 7SK with the Smn complex modulates snRNP production” by Changhe Ji, Jakob Bader, Pradhipa Ramanathan, Luisa Hennlein, Felix Meissner, Sibylle Jablonka, Matthias Mann, Utz Fischer, Michael Sendtner & Michael Briese. Nature Communications
Abstract
Interaction of 7SK with the Smn complex modulates snRNP production
Gene expression requires tight coordination of the molecular machineries that mediate transcription and splicing. While the interplay between transcription kinetics and spliceosome fidelity has been investigated before, less is known about mechanisms regulating the assembly of the spliceosomal machinery in response to transcription changes.
Here, we report an association of the Smn complex, which mediates spliceosomal snRNP biogenesis, with the 7SK complex involved in transcriptional regulation. We found that Smn interacts with the 7SK core components Larp7 and Mepce and specifically associates with 7SK subcomplexes containing hnRNP R. The association between Smn and 7SK complexes is enhanced upon transcriptional inhibition leading to reduced production of snRNPs.
Taken together, our findings reveal a functional association of Smn and 7SK complexes that is governed by global changes in transcription. Thus, in addition to its canonical nuclear role in transcriptional regulation, 7SK has cytosolic functions in fine-tuning spliceosome production according to transcriptional demand.
[ad_2]
Source link